HSPA8 (基因名), Heat shock cognate 71 kDa protein (蛋白名), hsp7c_horse.
产品名称:
Horse HSPA8/ Heat shock cognate 71 kDa protein ELISA Kit
货号:
-
商标:
EIAab®
监管等级:
别名:
Heat shock 70 kDa protein 8, HSC70
检测方法:
ELISA
特异性:
Natural and recombinant horse Heat shock cognate 71 kDa protein
样品类型:
Serum, plasma, tissue homogenates, cell culture supernates and other biological fluids
样品数据:
登录.
研究领域:
Epigenetics
通用注释
亚单元:
Identified in a IGF2BP1-dependent mRNP granule complex containing untranslated mRNAs. Interacts with PACRG. Interacts with HSPH1/HSP105. Interacts with IRAK1BP1 and BAG1. Interacts with DNAJC7. Interacts with DNAJB12 (via J domain). Interacts with DNAJB14 (via J domain). Interacts (via C-terminus) with the E3 ligase STUB1 forming a 210 kDa complex of one STUB1 and two HSPA8 molecules. Interacts with CITED1 (via N-terminus); the interaction suppresses the association of CITED1 to p300/CBP and Smad-mediated transcription transactivation. Component of the PRP19-CDC5L splicing complex composed of a core complex comprising a homotetramer of PRPF19, CDC5L, PLRG1 and BCAS2, and at least three less stably associated proteins CTNNBL1, CWC15 and HSPA8. Interacts with TRIM5. Part of a complex composed at least of ASCL2, EMSY, HCFC1, HSPA8, CCAR2, MATR3, MKI67, RBBP5, TUBB2A, WDR5 and ZNF335; this complex may have a histone H3-specific methyltransferase activity. Interacts with METTL21A. Following LPS binding, may form a complex with CXCR4, GDF5 and HSP90AA1. Interacts with PARK2. Interacts with FOXP3. Interacts with DNAJC9 (via J domain). Interacts with MLLT11. Interacts with RNF207. Interacts with DNAJC21. Interacts with DNAJB2. Interacts with TTC1 (via TPR repeats). Interacts with SGTA (via TPR repeats). Interacts with HSF1 (via transactivation domain). Interacts with HOPX, STUB1, HSP40, HSP901, BAG2 and BAG3.
功能:
Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, folding and transport of newly synthesized polypeptides, activation of proteolysis of misfolded proteins and the formation and dissociation of protein complexes. Plays a pivotal role in the protein quality control system, ensuring the correct folding of proteins, the re-folding of misfolded proteins and controlling the targeting of proteins for subsequent degradation. This is achieved through cycles of ATP binding, ATP hydrolysis and ADP release, mediated by co-chaperones. The co-chaperones have been shown to not only regulate different steps of the ATPase cycle of HSP70, but they also have an individual specificity such that one co-chaperone may promote folding of a substrate while another may promote degradation. The affinity of HSP70 for polypeptides is regulated by its nucleotide bound state. In the ATP-bound form, it has a low affinity for substrate proteins. However, upon hydrolysis of the ATP to ADP, it undergoes a conformational change that increases its affinity for substrate proteins. HSP70 goes through repeated cycles of ATP hydrolysis and nucleotide exchange, which permits cycles of substrate binding and release. The HSP70-associated co-chaperones are of three types: J-domain co-chaperones HSP40s (stimulate ATPase hydrolysis by HSP70), the nucleotide exchange factors (NEF) such as BAG1/2/3 (facilitate conversion of HSP70 from the ADP-bound to the ATP-bound state thereby promoting substrate release), and the TPR domain chaperones such as HOPX and STUB1. Acts as a repressor of transcriptional activation. Inhibits the transcriptional coactivator activity of CITED1 on Smad-mediated transcription. Component of the PRP19-CDC5L complex that forms an integral part of the spliceosome and is required for activating pre-mRNA splicing. May have a scaffolding role in the spliceosome assembly as it contacts all other components of the core complex. Binds bacterial lipopolysaccharide (LPS) and mediates LPS-induced inflammatory response, including TNF secretion. Participates in the ER-associated degradation (ERAD) quality control pathway in conjunction with J domain-containing co-chaperones and the E3 ligase STUB1.
亚细胞位置:
Cytoplasm
Melanosome
Nucleus
Nucleolus
Cell membrane
Localized in cytoplasmic mRNP granules containing untranslated mRNAs. Translocates rapidly from the cytoplasm to the nuclei, and especially to the nucleoli, upon heat shock (By similarity).
数据库链接
UniGene:
SMR:
STRING:
KEGG:
Pfam:
Uniprot:
该产品尚未在任何出版物中被引用。
[1].
马HSPA8ELISA试剂盒可以做多少个样本?
马HSPA8ELISA试剂盒分为2种规格,96孔和48孔。96孔的试剂盒,标曲和样本都做复孔的话,可以检测40个样本。96孔的试剂盒,标曲和样本都不做复孔的话,可以检测88个样本。
[2].
马HSPA8ELISA试剂盒使用视频?
马HSPA8ELISA试剂盒实验操作视频在以下网址中,对每一步的实验步骤都做了演示,方便实验员能更好地理解ELISA实验的过程。
https://www.eiaab.com.cn/lesson-tech/805.html
https://www.eiaab.com.cn/lesson-tech/805.html
[3].
马HSPA8ELISA试剂盒是放在-20℃冰箱保存吗?
EIAab的马HSPA8ELISA试剂盒,洗涤液、底物、终止液保存于4℃,其余试剂-20℃冰箱保存。
[4].
马HSPA8ELISA试剂盒原理?
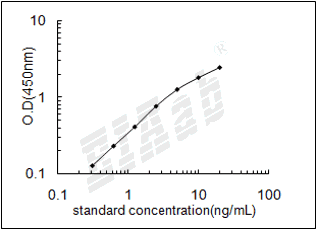
双抗体夹心法:用纯化的抗体包被微孔板,制成固相抗体,往包被有固相抗体的微孔中依次加入标准品或受检样本、生物素化抗体、HRP标记的亲和素,经过彻底洗涤后用底物TMB显色。用酶标仪在450nm波长下测定吸光度(OD值),计算样本浓度。
竞争法:用纯化的抗体包被微孔板,制成固相抗体,往包被有固相抗体的微孔中依次加入标准品或受检样本和生物素标记的目标分析物,受检标本中抗原与生物素标记抗原竞争结合有限的抗体。再加入HRP标记的亲和素,经过彻底洗涤后用底物TMB显色。用酶标仪在450nm波长下测定吸光度(OD值),计算样本浓度。
竞争法:用纯化的抗体包被微孔板,制成固相抗体,往包被有固相抗体的微孔中依次加入标准品或受检样本和生物素标记的目标分析物,受检标本中抗原与生物素标记抗原竞争结合有限的抗体。再加入HRP标记的亲和素,经过彻底洗涤后用底物TMB显色。用酶标仪在450nm波长下测定吸光度(OD值),计算样本浓度。
[5].
马HSPA8ELISA试剂盒中需要使用的样品量是多少?
夹心法100μL/孔,竞争法50μL/孔。如样本浓度过高时,应对样本进行稀释,以使稀释后的样本符合试剂盒的检测范围,计算时再乘以相应的稀释倍数。
[6].
如何分析马HSPA8ELISA试剂盒数据?
建议标准曲线,并计算样本浓度。对于elisa的曲线拟合,一般建议采用4参数曲线拟合,4参数曲线拟合通常更适合免疫分析。推荐使用专业软件进行曲线拟合,例如curve expert 1.3。根据样本的OD值由标曲查出相应的浓度,再乘以稀释倍数;或用标准物的浓度与OD值计算出标曲的回归方程式,将样本的OD值代入方程式,计算出样本浓度,再乘以稀释倍数,即为样本的实际浓度。以下链接是curve expert 1.3软件拟合曲线的方法。
https://www.eiaab.com.cn/news/502/
https://www.eiaab.com.cn/news/502/
[7].
马HSPA8ELISA试剂盒中是否包含人和动物的副产物,是否包含感染的或者传染性原料如HIV等?
除了抗体和稀释液中的BSA,不含其它人和动物的副产物,也不含感染材料。
[8].
收集马HSPA8ELISA试剂盒血浆样本,用什么作为抗凝剂?
一般建议用EDTA和肝素作为抗凝剂。
[9].
马HSPA8ELISA试剂盒酶标板可以拆成几部分?拆的时候是否需要避光,无菌?
马HSPA8ELISA试剂盒酶标板是8×12孔条,可拆卸,板子可以拆成12条,注意避免孔污染,不需要避光和无菌。暂时不用的板子,放回原来装的袋子里,密封保存。
[10].
马HSPA8ELISA试剂盒样本如何保存?
尽量检测新鲜样本。若无新鲜样本,则4℃保存1周,-20℃保存1个月,-80℃保存2个月。
反馈墙
评论数 : 0
所有用户
所有用户
默认排序
默认排序
最近
早期
目前还没有评论。



通知
规格
数量
单价 (¥)
小计 1 (¥)
小计 2:
¥

规格
数量
单价 (¥)




 验证序列:
验证序列:




 折扣:
折扣: